|
 I am a cognitive neuroscientist, specializing in the development of novel analytic methods for large-scale
human neuroimaging datasets. I have a particular interest in using the parallel lines of
evidence provided by multimodal datasets, to get a better understanding of
the neural correlates (and ultimately mechanisms) of human cognitive functions, both in the
young, healthy brain, and in older, degenerating ones. I am a cognitive neuroscientist, specializing in the development of novel analytic methods for large-scale
human neuroimaging datasets. I have a particular interest in using the parallel lines of
evidence provided by multimodal datasets, to get a better understanding of
the neural correlates (and ultimately mechanisms) of human cognitive functions, both in the
young, healthy brain, and in older, degenerating ones.
The methods I use include:
 Pupillometry measures the size and gaze position of a subject's pupils. Gaze position can be used to estimate what part of the visual field a person is attending to, while pupil diameter is a useful proxy measure for a person's level of arousal. I use this method to investigate the role of norepinephrine in cognition. Pupillometry measures the size and gaze position of a subject's pupils. Gaze position can be used to estimate what part of the visual field a person is attending to, while pupil diameter is a useful proxy measure for a person's level of arousal. I use this method to investigate the role of norepinephrine in cognition.
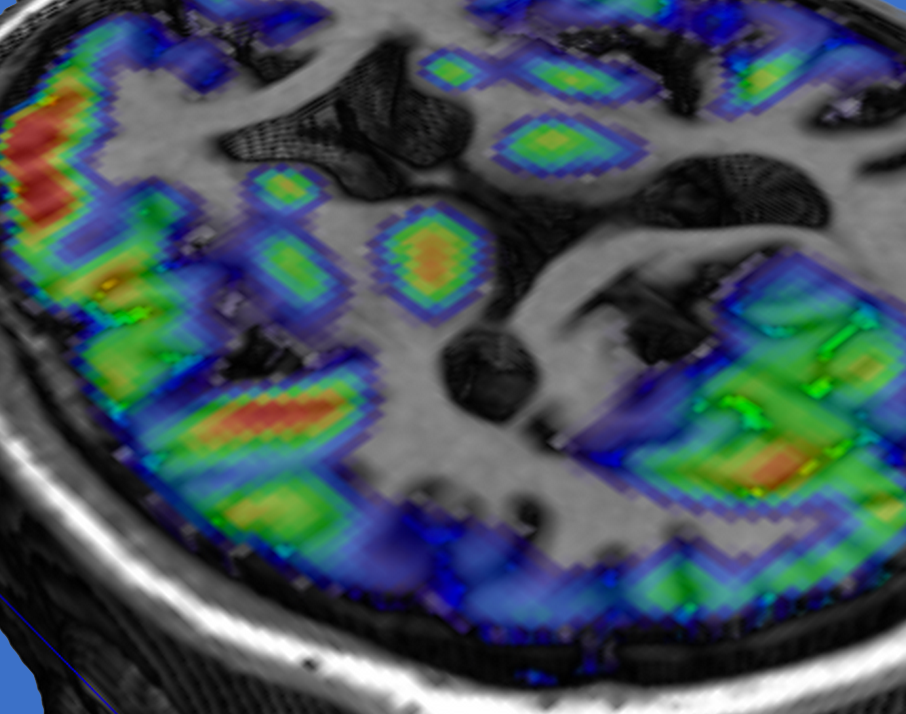
 Cortical thickness estimation involves the use of T1-weighted anatomical
images to estimate how thick the cortical sheet is at different locations. This
method allows us to get an idea, for instance, how brain structure differs for different groups of
people, or how it degenerates in conditions such as Alzheimer's disease. Cortical thickness estimation involves the use of T1-weighted anatomical
images to estimate how thick the cortical sheet is at different locations. This
method allows us to get an idea, for instance, how brain structure differs for different groups of
people, or how it degenerates in conditions such as Alzheimer's disease.
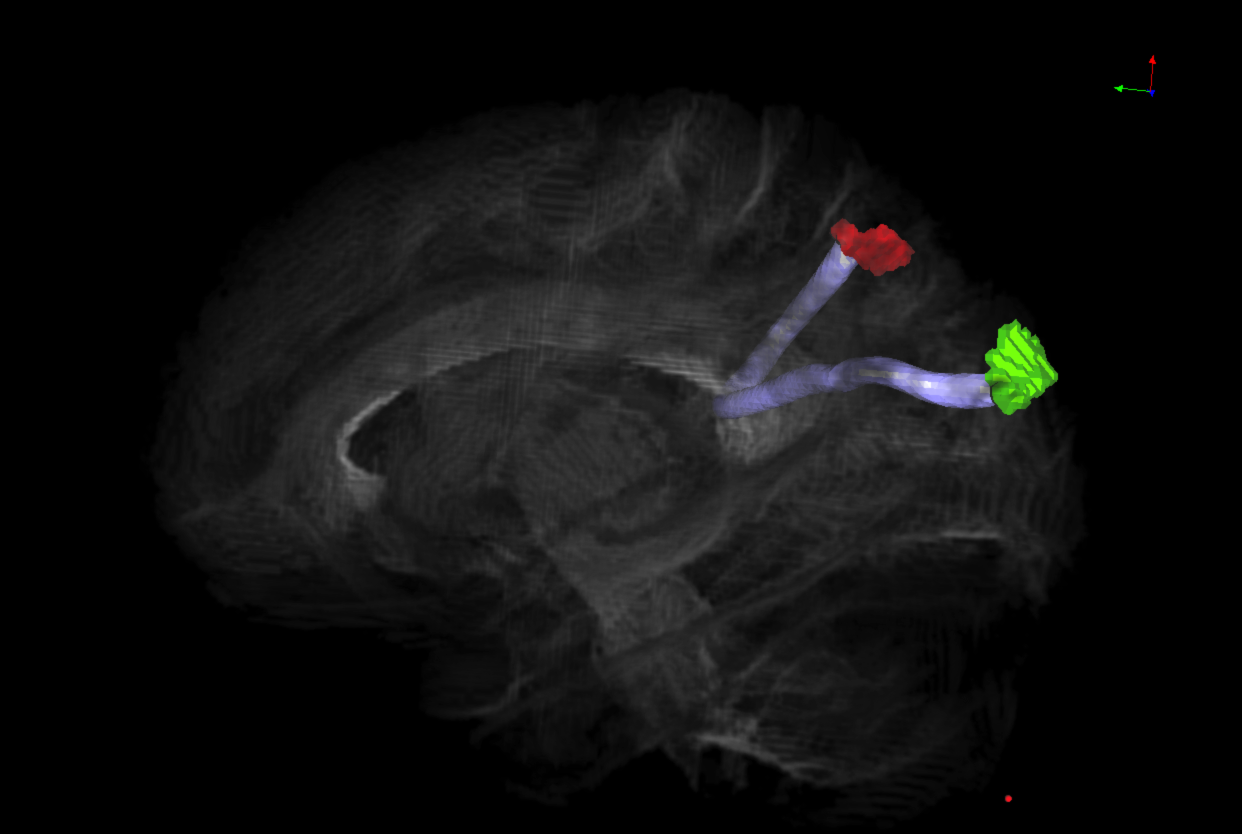
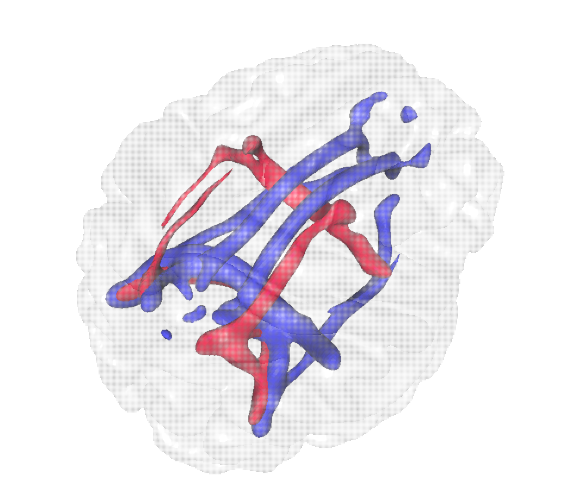 Diffusion-weighted imaging (DWI) is a
type of magnetic resonance imaging (MRI) acquisition technique which allows us to
estimate the diffusion of molecules in the brain. DWI is used to perform probabilistic
tractography, which allows us to probe the geometry of myelinated axons, which form the "wiring"
of brain networks. Diffusion-weighted imaging (DWI) is a
type of magnetic resonance imaging (MRI) acquisition technique which allows us to
estimate the diffusion of molecules in the brain. DWI is used to perform probabilistic
tractography, which allows us to probe the geometry of myelinated axons, which form the "wiring"
of brain networks.
 Resting-state functional MRI (rs-fMRI)
is another MRI-based technique which measures blood oxygenation (BOLD) in the brains of subjects who
are not given explicit task demands. Rs-fMRI allows us to estimate "instrinsic" covariance in brain
activity, which provides (indirect) information about the brain's ability to communicate and process
information. Resting-state functional MRI (rs-fMRI)
is another MRI-based technique which measures blood oxygenation (BOLD) in the brains of subjects who
are not given explicit task demands. Rs-fMRI allows us to estimate "instrinsic" covariance in brain
activity, which provides (indirect) information about the brain's ability to communicate and process
information.
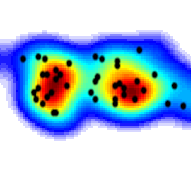 Meta-analysis is the statistical analysis
of databases which collate a large number of published scientific findings. I have used meta-analysis to
study whole-brain connectivity using macaque monkey tract tracing, compiled in the CoCoMac database.
I have also used a method called activation likelihood estimation (ALE) to study fMRI activation
drawn from thousands of cognitive paradigms, collated in the BrainMap database. Meta-analysis is the statistical analysis
of databases which collate a large number of published scientific findings. I have used meta-analysis to
study whole-brain connectivity using macaque monkey tract tracing, compiled in the CoCoMac database.
I have also used a method called activation likelihood estimation (ALE) to study fMRI activation
drawn from thousands of cognitive paradigms, collated in the BrainMap database.
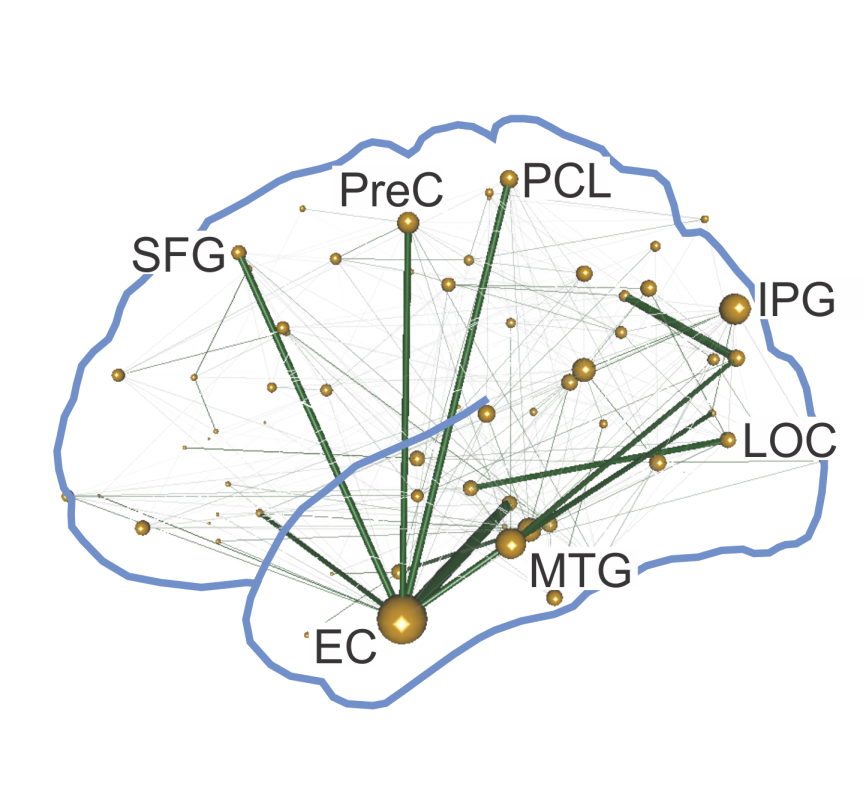 Network analysis is the application of
mathematical tools to the analysis of a network's organization and topology. I have used such approaches
to investigate the hierarchical organization of the macaque brain, based upon laminar profiles of axonal
projections between brain regions. I have also used graph theoretical approaches to evaluate
the importance of individual brain regions to the overall connectivity of the network. Network analysis is the application of
mathematical tools to the analysis of a network's organization and topology. I have used such approaches
to investigate the hierarchical organization of the macaque brain, based upon laminar profiles of axonal
projections between brain regions. I have also used graph theoretical approaches to evaluate
the importance of individual brain regions to the overall connectivity of the network.
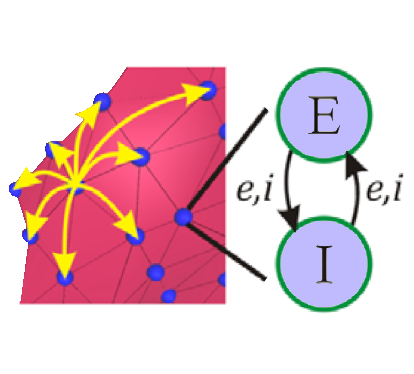 Mean field modelling (MFM) simulates brain activity
by treating large populations of neurons as single units, and generating mean firing rates for each unit.
MFM allows us to simulate the brain signals that we observe with empirical methods such as fMRI, electroencephalography
(EEG), and magnetoencephalography (MEG). Activity can spread both locally (along the cortical "field") and
through long-range axonal connections. Mean field modelling (MFM) simulates brain activity
by treating large populations of neurons as single units, and generating mean firing rates for each unit.
MFM allows us to simulate the brain signals that we observe with empirical methods such as fMRI, electroencephalography
(EEG), and magnetoencephalography (MEG). Activity can spread both locally (along the cortical "field") and
through long-range axonal connections.
|